ML-SIM
Read more or try it out now below
The preprint paper describing this method is referenced below.
You can see some examples in the bottom of the page.
Our new manuscript, ML-SIM, is now on arXiv: https://t.co/BQo1VH4NiN
— Charles Nicklas Christensen (@charlesnchr) March 27, 2020
Sadly I won't get to present the work at @FOMconferences this year (#COVID19 😑), but at least I am happy to provide some non-COVID reading material for those interested. #ML_SIM #group_laser pic.twitter.com/Z9XrcOscQy
Get the source code now at GitHub.
Want
to see how it works?
Give ML-SIM a try with
your own images - simply drop them in the box below. You can also use the ML-SIM Test Images that consist of simulated images and data from two distinct microscopes (SLM-based and interferometric, respectively).
Note that the currently served model is trained on 9-frame SIM stacks of 512x512 resolution.
Examples
See the difference between reconstructions from
ML-SIM and standard wide-field microscopy.
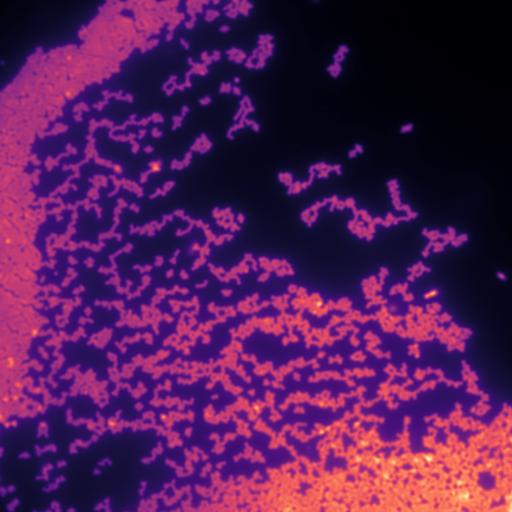
Tried the new machine-learning #superresolution structured illumination #microscopy reconstruction algorithm https://t.co/NgeeUhaTGX. Nephrin staining of a kidney comparing widefield and @zeiss_micro + ML-SIM algorithms. Thanks @palankarr for pointing out! https://t.co/9AtHprz4wg pic.twitter.com/WHh1pgzu6z
— Florian Siegerist (@Siegerist) March 29, 2020
Test of ML-SIM posted on Twitter.
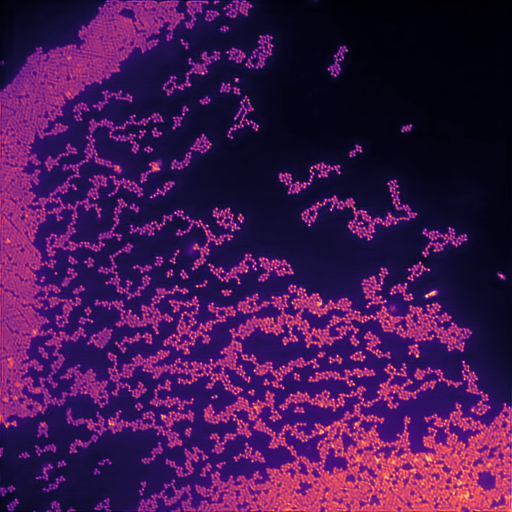
Calibration structured illumination image of beads.
Test image with simulated diffraction and illumination patterns.
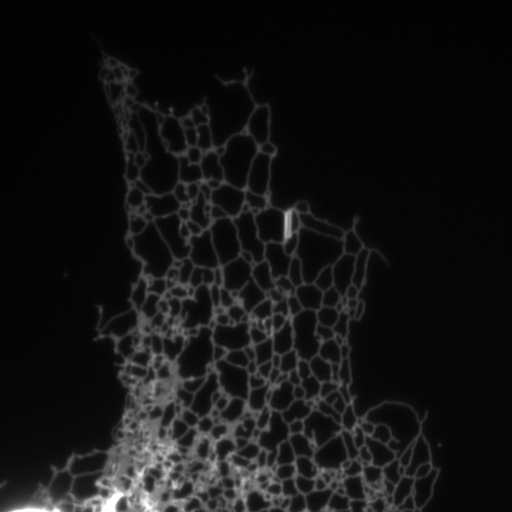
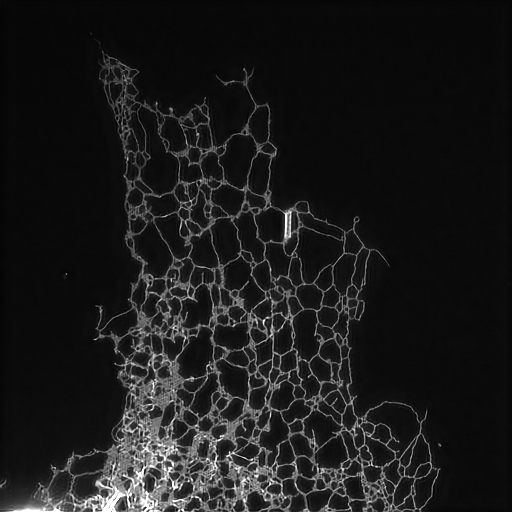
Structured illumination image of endoplasmic reticulum.